The biological process, defined as translation, which allows the expression of mRNA in proteins, occurs through specific macromolecular complexes: the ribosomes. (Fig. 1)

Ribosomes discovered by the biologist George Emil Palade through the microscope, are characterized by two macromolecular ribosomal RNA (rRNA) and proteins. Ribosomal proteins and RNA are aggregated through non-covalent interaction, primarily by hydrogen bonds. These structures can be found in the cytoplasm of all cells, both prokaryotic and eukaryotic. In prokaryotes, these organelles are free, as there is no endoplasmic reticulum that can bind them.
Structures: prokaryotes and eukaryotes
The two asymmetric subunits of the ribosomes, observed under a microscope, seem to be structurally very similar. Meanwhile the largest fraction has a boat shape with a more pointed end, the smallest fraction is formed by a head and a body tied by a thinner neck. Bacterial ribosomes with a mass of about 2700 kDa and a diameter of 20 nm, have a sedimentation coefficient of 70S (Fig.2) and the two fractions that make up the complex have respective values of 30S and 50S. In detail, the 30S subunit consists of 21 different proteins and a molecule of rRNA with a coefficient of 16S. Instead, the 50S fraction is formed by 34 proteins and two RNA molecules (5S and 23S).
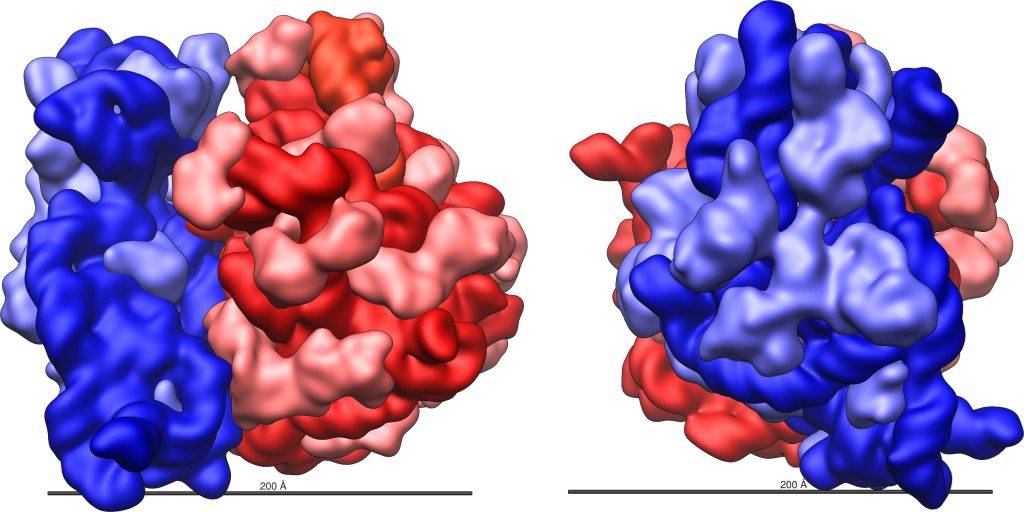
Unlike eukaryotic ribosomes which have a mass of 4000kDa and show a total sedimentation coefficient of 80S and two subunits of 60S (45 proteins and 3 rRNA molecules) and 40S respectively (33 proteins and one rRNA molecule 18S). It should be noted that the sedimentation coefficient, indicated by the letter S, in Svedberg units, refers to the sedimentation rate in ultra-speed centrifugation. Factors such as molecular weight, size, weight, and shape affect the parameter. For these reasons, the sum of the two subunits (30S + 50S) does not correspond to the total coefficient of 70S.
Structure: relationship with function
A flat region (platform) in the slight curvature of the minor subunit forms a groove in correspondence with the major subunit depression. The furrow between the two subunits will receive the mRNA during the translation. In the largest fraction of the ribosome there are three binding sites for tRNA molecules:
- A: site of attack, where tRNA anticodons binds to mRNA codon, placing its amino acid next to the polypeptide chain that is forming.
- P (polypeptide site): tRNA yields its amino acid to the forming chain at the condensation site.
- E (site Exit): the tRNA after giving up its amino acid, is placed in the site of detachment, before leaving the ribosome and restart the cycle.
Function: Formation of the start-up complex
As you can well understand, ribosomes play a fundamental role in protein synthesis. We recall the phases of prokaryotic and eukaryotic translation, which follow one another schematically :

At the beginning the rRNA present in the minor subunit of the ribosome binds to the binding site located near end 5′ on the mRNA. The bond between the ribosome and the mRNA is facilitated, in prokaryotes, by the Shine-Dalgarno sequences, bases present between 5 and 10 nucleotides before the starting codon. Meanwhile the complementary tRNA anticodons, loaded with formed methionine at the level of nitrogen (fMet) or methionine in the case of eukaryotes, binds to the initial AUG codon of the mRNA.
The major fraction then binds to the starting complex consisting of the loaded tRNA of the amino acid together with the minor subunit and mRNA. The tRNA with fMet/Met passes the site P and in site A the second codon of the mRNA is placed. Each tRNA is specific to a single amino acid, the free hydroxyl in 3 of the tRNA acceptor arm is the hook point of the amino acid.
In the first phase of translation, the main difference between prokaryotes and eukaryotes concerns initiation factors. The initiation factors of prokaryotes allow the binding of all components, perform activities for the preparation of the ribosome: IF1 removes the subunits 30S and 50S, IF2 promotes the binding of the fMet-tRNA complex to the 30S ribosome and IF3 binds the mRNA to the 30S subunit. In eukaryotes, we must remember that the mRNA has a CAP and a hairpin to deal with multiple initiation factors (EIF4F, EIF2, EIF2B, EIF3, EIF6, EIF5, EIF1, and EIF1A) to facilitate the ribosome in its work.
Function: Elongation
In site P, lost the link with the Formylmethionine, binds the second amino acid loaded on the tRNA located in site A. The major fraction of the ribosome catalyzes the peptide bond between the two amino acids, so rRNA works as a ribozyme. From site P, the amino acid-free tRNA passes to site E and comes off. The ribosome proceeds in directions 5′-3′ and a codon, so that the tRNA-polypeptide complex is located at the site P. In prokaryotes, the factors EF-1A, EF-1B, and EF-2 intervene in this phase of elongation. EF-1A brings the next tRNA to site A of the ribosome, EF-1B acts as a nucleotide exchange factor replacing the resulting GDP molecule with a new GTP and the last EF-2 factor averaging the translocation. While in eukaryotes, eEF-1 brings the next amino-acyl tRNA to site A of the ribosome and eEF-2 media intervenes in the translocation.
Function: Termination
The last phase occurs when the enzyme encounters stop codons (UAA, UAG, and UGA). No tRNA binds to stop codons but release factors. In RF1 prokaryotes it recognizes the codons UUA and UAG, while RF2 UAA and UGA and their ribosome binding is assisted by RF3 (GTP-dependent protein). In eukaryotes there are two release factors, eRF1 recognizes the three termination codons and eRF3 supports the detachment of the polypeptide from the ribosome.
Original article by: Veronica Nerino – Translated by Luigi Gallucci