There is a part of human DNA whose role is not yet well known, which is why it has been given the name of “junk” DNA. Only recently a study showed that “junk” DNA is not as inert as it is thought, on the contrary it would seem to contribute to the accumulation of mutations that may be at the basis of cancer development.
What is “junk” DNA?
In bacteria, the regions of the genome coding for proteins generally occupy 88%. The remaining 12% consists largely of non-coding genes and regulatory sequences. In eukaryotes, however, the situation is different. The amount of coding DNA is usually a much smaller fraction of the total: the human genome, for example, contains 3.2 billion base pairs of which only between 1% and 2% represent coding DNA. The remaining 98-99% consists of the so-called “non-coding DNA” (ncDNA).
Some of the non-coding DNA is transcribed into functional RNA molecules (eg transfer RNA, microRNA, piRNA, ribosomal RNA, and regulatory RNA).
Other functional regions of the non-coding DNA fraction include regulatory sequences that control the expression of some genes, binding sites for proteins or enzymes, origins of DNA replication and structural chromosomal elements (centromeres and telomeres).
Some regions, on the other hand, appear to be apparently non-functional such as introns, pseudogenes, repetitive sequences and fragments of transposons and viruses. These regions occupy most of the genome of many eukaryotes and have been dubbed junk DNA.
This term which became popular in the 1960s initially referred to all non-coding DNA. Today, however, it refers to a portion of non-coding DNA that does not have a defined function in development, physiology or some other capacity at the level of the organism.
Stalled DNA replication
As with coding DNA, “junk” DNA is also subjected to the replication process. Accurate replication of chromosomal DNA, including that of junk DNA, is essential to maintain genomic stability.
Under normal conditions, the synthesis of the leading strand is coupled to the unwinding of double-stranded DNA (dsDNA) into single-stranded DNA (ssDNA). In such conditions the synthesis reaches the maximum speed. However, polymerase synthesis activity can stall, and therefore stop, due to a number of factors, including DNA damage, DNA-bound proteins, collisions with transcriptional machinery, RNA-DNA hybrids (R- loops), stressors and limiting dNTPs (deoxynucleotides triphosphate).

Despite the block, however, the helicase can still continue to carry out the dsDNA at a reduced speed. This situation is called helicase-polymerase decoupling and is capable of activating a series of repair mechanisms similar to those induced by strand breakage. All this can cause genetic instability which makes it more susceptible to the accumulation of mutations.
In addition to the exogenous factors described above, some sequences of the DNA itself can cause stalling. These sequences do not necessarily have to be located in the coding portion of the DNA, but also in the non-coding one and more particularly in the “junk” DNA itself.
What sequences can affect replication?
To date, most of the evidence on how DNA affects its own replication comes from highly repetitive sequence studies. It is a class of DNA sequences in which two or more nucleotides are repeated next to each other. One of the characteristics of these repetitions is that they can expand from one generation to the next, increasing the probability that the function of a gene can be interrupted, causing a series of pathologies.
The best known mechanism capable of causing the expansion of repetitive sequences is the so-called “replication slippage“. When there are areas with long repetitions in the DNA, the polymerase encounters the same sequence several times. This can confuse the polymerase: the filament slips and starts the synthesis of an already replicated sequence.
Another feature of these sequences is that, once the DNA has been separated into single-stranded DNA, they have a propensity to fold into “unusual” secondary structures. Some examples are:
• hairpins: double-stranded structures that are formed by the classic Watson and Crick type pairing between the bases of two complementary sequences adjacent and inversely arranged;
• G-quadruplex: four-stranded DNA structures formed through hydrogen bonds between guanine-rich sequences;
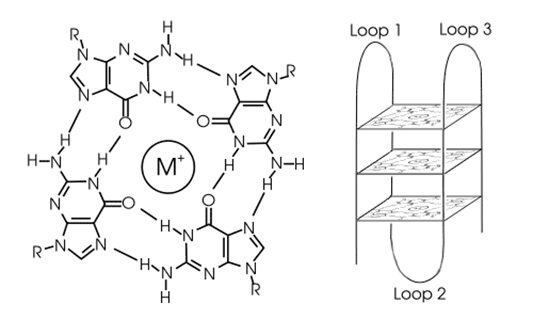
• i-motifs: four-stranded DNA structures rich in cytosine, similar to G-quadruplexes;
• triplex: triple-stranded DNA structures. The double helix is formed through the coupling of Watson-Crick bases, while the third strand is coupled to the double helix.
These structures create a footprint that could interfere with the complex of proteins involved in replication (replisome), slowing it down or even blocking it.
“Junk” DNA can cause replication to stop
A recent study conducted at the Genome Replication lab of the Institute of Cancer Research in London, published in Nature Communications showed that the repetitive patterns of DNA, abundantly present in “junk” DNA, are able to block replication, increasing the risk of mistakes that can be one of the first causes of cancer.
In this study, the scientists reconstructed what happens to the replisome when it encounters repetitive DNA sequences. In particular, they reconstituted the entire DNA replication process in vitro using a replisome isolated and purified from yeast proteins. The research group conducted all the experiments in the absence of other protein components capable of blocking the replication process, so as to make it dependent on the DNA sequence alone.
After testing a wide range of repetitive mono, di and trinucleotides, they observed that these sequences were able to induce replication stall. This ability was more related to the type of secondary DNA structures that were formed. In particular, it was found that the polymerase was intrinsically capable of continuing the synthesis despite the formation of the double helix structures (hairpins). On the contrary, the formation of the quadruple helix structures (G-quadruplex and i-motifs) induced the block of the polymerase, with the consequent uncoupling of the helicase-polymerase and was solved only by the same repair mechanisms induced by the DNA lesions.
Conclusions
This study showed that the repetitive sequences abundantly present in “junk” DNA are able to slow down or block DNA replication, triggering repair mechanisms and therefore contributing to genetic instability. This could cause the development of mutations underlying the onset of cancer. For the first time, therefore, one of the still under-explored aspects of “junk” DNA has been investigated, suggesting that this could play an important and potentially harmful role in cells.
Original article “Il DNA “spazzatura” e il suo ruolo nel cancro” written by Ivana Bello
Translation by Giovanna Spinosa
Sources
- https://www.icr.ac.uk/news-archive/junk-dna-could-lead-to-cancer-by-stopping-copying-of-dna;
- https://en.wikipedia.org/wiki/Non-coding_DNA;
- Casas-Delucchi, C.S., Daza-Martin, M., Williams, S.L. et al. The mechanism of replication stalling and recovery within repetitive DNA. Nat Commun 13, 3953 (2022). https://doi.org/10.1038/s41467-022-31657-x.